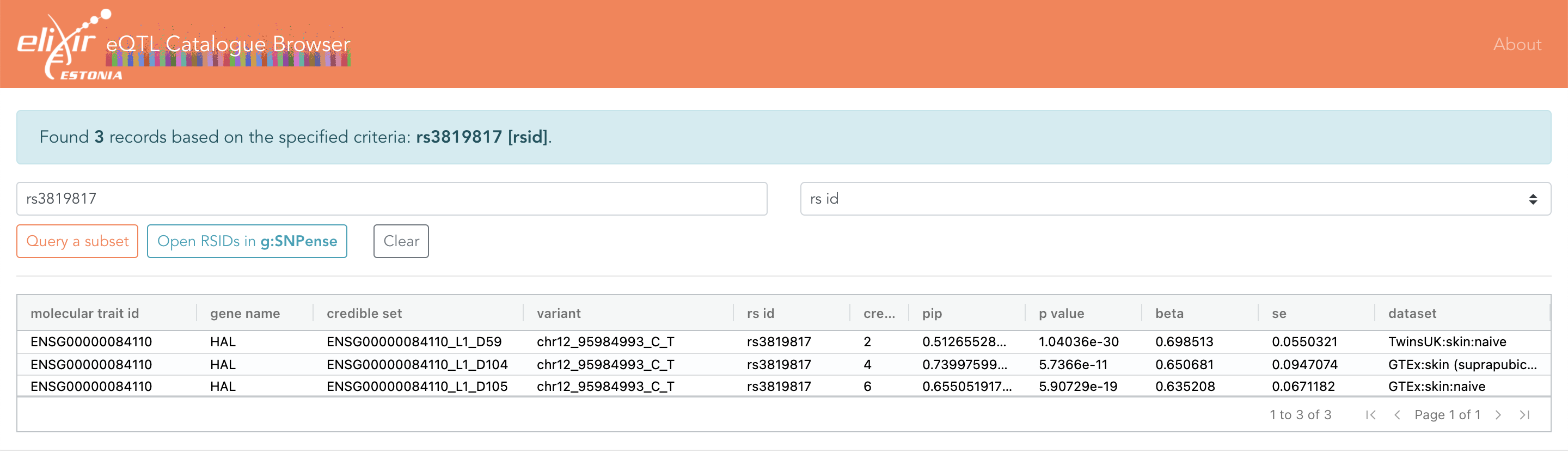
Eqtl Catalogue
Eqtl Catalogue - Rapid querying, colocalization, and plotting of summary stats from the eqtl catalogue. The eqtl catalogue is an open database of uniformly processed human molecular quantitative trait loci (qtls). The eqtl catalogue is a compendium of uniformly processed eqtls from 21 studies, covering various cell types and tissues. 29 rows the eqtl catalogue is a database of expression quantitative trait loci (eqtl). The eqtl catalogue provides uniform processing of 21 eqtl studies, allowing the identification of highly reproducible eqtls affecting whole gene and transcript expression levels. The eqtl catalogue is a new project aimed at accelerating the functional interpretation of human gwas studies and the annotation of common variants through a comprehensive collection of. Eqtl catalogue currently contains >100 datasets from 20 different studies. It enables the integration, fine mapping and. The four primary workflows are: All data analysis for the eqtl catalogue is performed using reproducible and containerised nextflow workflows. The eqtl catalogue is a compendium of uniformly processed eqtls from 21 studies, covering various cell types and tissues. The four primary workflows are: You can access the results via ftp, api or open targets genetics portal. All data analysis for the eqtl catalogue is performed using reproducible and containerised nextflow workflows. 29 rows the eqtl catalogue is a database of expression quantitative trait loci (eqtl). We are continuously updating the resource to further. The eqtl catalogue provides uniform processing of 21 eqtl studies, allowing the identification of highly reproducible eqtls affecting whole gene and transcript expression levels. It enables the integration, fine mapping and. Eqtl catalogue currently contains >100 datasets from 20 different studies. Currently hosting eqtl and sqtl data from 16 different studies from the ebi eqtl catalogue It enables the integration, fine mapping and. The eqtl catalogue is a new project aimed at accelerating the functional interpretation of human gwas studies and the annotation of common variants through a comprehensive collection of. Eqtl catalogue currently contains >100 datasets from 20 different studies. You can access the results via ftp, api or open targets genetics portal. Rapid querying,. 29 rows the eqtl catalogue is a database of expression quantitative trait loci (eqtl). The eqtl catalogue provides uniform processing of 21 eqtl studies, allowing the identification of highly reproducible eqtls affecting whole gene and transcript expression levels. We are continuously updating the resource to further. The eqtl catalogue is a compendium of uniformly processed eqtls from 21 studies, covering. The eqtl catalogue provides uniform processing of 21 eqtl studies, allowing the identification of highly reproducible eqtls affecting whole gene and transcript expression levels. Currently hosting eqtl and sqtl data from 16 different studies from the ebi eqtl catalogue 29 rows the eqtl catalogue is a database of expression quantitative trait loci (eqtl). Rapid querying, colocalization, and plotting of summary. The eqtl catalogue is a compendium of uniformly processed eqtls from 21 studies, covering various cell types and tissues. You can access the results via ftp, api or open targets genetics portal. The eqtl catalogue provides uniform processing of 21 eqtl studies, allowing the identification of highly reproducible eqtls affecting whole gene and transcript expression levels. The four primary workflows. The eqtl catalogue is an open database of uniformly processed human molecular quantitative trait loci (qtls). It enables the integration, fine mapping and. The four primary workflows are: Rapid querying, colocalization, and plotting of summary stats from the eqtl catalogue. The eqtl catalogue is a new project aimed at accelerating the functional interpretation of human gwas studies and the annotation. The four primary workflows are: The eqtl catalogue is a new project aimed at accelerating the functional interpretation of human gwas studies and the annotation of common variants through a comprehensive collection of. Rapid querying, colocalization, and plotting of summary stats from the eqtl catalogue. You can access the results via ftp, api or open targets genetics portal. All data. Eqtl catalogue currently contains >100 datasets from 20 different studies. You can access the results via ftp, api or open targets genetics portal. We are continuously updating the resource to further. 29 rows the eqtl catalogue is a database of expression quantitative trait loci (eqtl). The eqtl catalogue is a new project aimed at accelerating the functional interpretation of human. It enables the integration, fine mapping and. We are continuously updating the resource to further. The eqtl catalogue is an open database of uniformly processed human molecular quantitative trait loci (qtls). Currently hosting eqtl and sqtl data from 16 different studies from the ebi eqtl catalogue All data analysis for the eqtl catalogue is performed using reproducible and containerised nextflow. All data analysis for the eqtl catalogue is performed using reproducible and containerised nextflow workflows. Currently hosting eqtl and sqtl data from 16 different studies from the ebi eqtl catalogue The eqtl catalogue is a compendium of uniformly processed eqtls from 21 studies, covering various cell types and tissues. The four primary workflows are: You can access the results via. It enables the integration, fine mapping and. 29 rows the eqtl catalogue is a database of expression quantitative trait loci (eqtl). The eqtl catalogue is a compendium of uniformly processed eqtls from 21 studies, covering various cell types and tissues. The four primary workflows are: You can access the results via ftp, api or open targets genetics portal. It enables the integration, fine mapping and. Eqtl catalogue currently contains >100 datasets from 20 different studies. All data analysis for the eqtl catalogue is performed using reproducible and containerised nextflow workflows. The eqtl catalogue provides uniform processing of 21 eqtl studies, allowing the identification of highly reproducible eqtls affecting whole gene and transcript expression levels. We are continuously updating the resource to further. Currently hosting eqtl and sqtl data from 16 different studies from the ebi eqtl catalogue The eqtl catalogue is an open database of uniformly processed human molecular quantitative trait loci (qtls). The four primary workflows are: You can access the results via ftp, api or open targets genetics portal. 29 rows the eqtl catalogue is a database of expression quantitative trait loci (eqtl).Building the eQTL Catalogue a Q&A with Kaur Alasoo
Overview of studies and samples included in the eQTL Catalogue a
(PDF) eQTL Catalogue 2023 New datasets, X chromosome QTLs, and
Overview of the eQTL Catalogue database a, A highlevel representation
Building the eQTL Catalogue a Q&A with Kaur Alasoo
Overview of the eQTL Catalogue database a, A highlevel representation
(PDF) eQTL Catalogue a compendium of uniformly processed human gene
GitHub eQTLCatalogue/eQTLCatalogueresources
Overview of the datasets included in the eQTL Catalogue. (A
eQTL Catalogue a new resource for gene expression and splicing QTLs
The Eqtl Catalogue Is A New Project Aimed At Accelerating The Functional Interpretation Of Human Gwas Studies And The Annotation Of Common Variants Through A Comprehensive Collection Of.
Rapid Querying, Colocalization, And Plotting Of Summary Stats From The Eqtl Catalogue.
The Eqtl Catalogue Is A Compendium Of Uniformly Processed Eqtls From 21 Studies, Covering Various Cell Types And Tissues.
Related Post: